I am interested in how bacteria respond to stresses such as iron limitation, antibiotics, and barriers to motility. The systems used by bacteria to overcome these stresses are required for growth during infection and are potential targets for therapy. We investigate the function of proteins involved in stress response through a combination of structure determination, genetic phenotypes, and biochemical assays. Below are descriptions of four ongoing projects with some reference to related publications.
(a) Iron and oxidative stress
Within the cell, the labile iron pool is believed to not bound to proteins as cofactors but chelated in the ferrous form by metabolites. This iron may be oxidized by ferritin and stored as a ferric-oxide within the ferritin nanocage. Iron movement from the labile pool to ferritin is hypothesized to be in response to oxidative stress. Our aim is to show the conditions under which ferroxidation by ferritin is important for cell survival.
Pfaffen, S., Bradley, J.M., Abdulqadir, R., Firme, M.R., Moore, G.R, Le Brun, N.E. and Murphy, M.E.P. (2015) A diatom ferritin optimized for iron oxidation but not iron storage. J. Biol. Chem. 290: 28416-27. (doi: 10.1074/jbc.M115.669713)
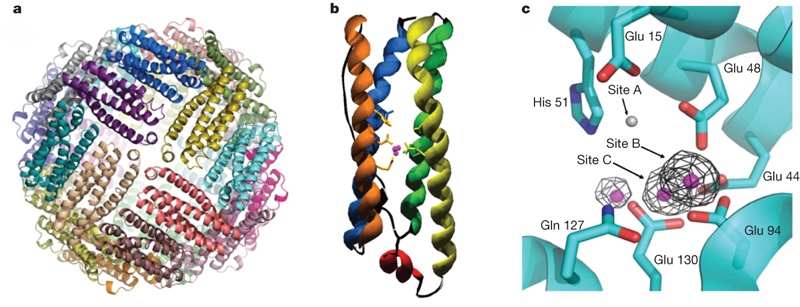
(b) Helical cell shape of epsilon-proteobacteria
The pathogens Campylobacter jejuni and Helicobacter pylori have helical cell shape used to burrow into mucosal layers of gastrointestinal track. Bacterial cell shape is defined by the structure of the peptidoglycan layer and helical bacteria have enzymes that modify this layer and are required for helical morphology. We are investigating the structure and function of these cell shape determining enzymes. This project is a collaboration with Dr. Tanner (Chemistry), Dr. Gaynor (Microbiology and Immunology) and Dr. Salma (FHCRC, Seattle).
Liu, Y., Frirdich, E., Taylor, J.A., Chan, A.C.K., Blair, K.M., Vermeulen, J., Ha, R., Murphy, M.E.P., Salama, N.R., Gaynor, E.C. & Tanner, M.E. (2016) A bacterial cell shape-determining inhibitor. ACS Chem. & Biol. 11: 981-991. DOI: 10.1021/acschembio.5b01039.
Chan, A.C.K., Blair, K.M., Liu, Y., Frirdich, E., Gaynor, E.C., Tanner, M.E., Salama, N.R. & Murphy. M.E.P. (2015) Helical shape of Helicobacter pylori requires an atypical glutamine as a zinc ligand in the carboxypeptidase Csd4. J. Biol. Chem. 290, 3622-38.
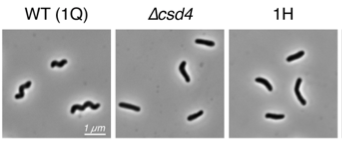
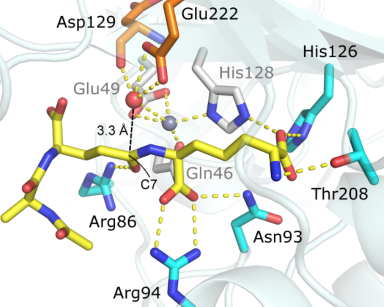
Above, images of of wild-type H. pylori cells with a helcial shape and mutants with either deletion of the csd4 gene or a point mutation at Glu49. Below, the structure of a synthetic tripeptide mimetic of substrate peptidoglycan bound to Csd4.
(c) Antibiotic resistance by Burkholderia
Burkholderia cenocepacia complex is a group of bacteria that infect the lungs of cystic fibrosis patients. These bacteria are notoriously resistant to clinically available antibiotics. We are investigating two mechanisms responsible to broad antibiotic resistance by these bacteria. The first is the secretion of bacterial lipocalins which potentially bind antibiotics and the second is modification of lipid A (a collaboration with Dr. Valvano at Queen’s University Belfast).
El-Halfawy, O.M., Klett, J., Ingram, R.J., Loutet, S.A., Murphy, M.E.P., Martín-Santamaría, S., and Valvano, M.A. (2017) Antibiotic capture by bacterial lipocalins uncovers an extracellular mechanism of intrinsic antibiotic resistance. mBio 8: e00225-17.
(d) Staphylococcal response to iron limitation
The bacterial pathogen Staphylococcus aureus uses iron for metabolism and to counter oxidative stress. During invasive infection, iron is acquired by S. aureus from host sources including heme from hemoglobin. Also, the pathogen adapts to iron limiting growth conditions by repressing iron-requiring pathways such as the TCA cycle and oxidative phosphorylation. We are investigating siderophore and heme-iron based uptake systems used by S. aureus to acquire iron from the human host environment. For example, IsdB is expressed on the bacterial cell surface and binds hemoglobin and extracts heme for use by S. aureus. Staphyloferrin B is a siderophore produced by S. aureus by the enzymes expressed from the sbn locus. We interested in understanding how staphyloferrin B is produced when S. aureus metabolism is operating under iron limitation. We are collaborating with Dr. Heinrichs (U. Western Ontario)
Verstraete M.M., Perez-Borrajero C., Brown K.L., Heinrichs D.E., & Murphy M.E.P. (2018) SbnI is a free serine kinase that generates O-phospho-L-serine for staphyloferrin B biosynthesis in Staphylococcus aureus. J. Biol. Chem. 2018 Feb 26. pii: jbc.RA118.001875. doi: 10.1074/jbc.RA118.001875.
Bowden, C.M.F., Chan, A.C.K., Li, E.J.W., Arrieta, A.L., Eltis, L.D. & Murphy, M.E.P. (2018) Structure-function analyses reveal key features in Staphylococcus aureus IsdB-associated unfolding of the heme-binding pocket of human hemoglobin. J. Biol. Chem. 293, 177-190.
Kobylarz, M.J., Heieis, G.A., Loutet, S.A. & Murphy, M.E.P. (2017) Iron Uptake Oxidoreductase (IruO) Uses a Flavin Adenine Dinucleotide Semiquinone Intermediate for Iron-Siderophore Reduction. ACS Chem. & Biol. 12: 1778-86. doi: 10.1021/acschembio.7b00203.
Kobylarz, M.J., Grigg, J.C., Takayama, S.J., Rai, D.Y., Heinrichs, D.E., and Murphy, M.E.P. (2014) Synthesis of L-2,3-diaminopropionic acid, a siderophore and antibiotic precursor. Chem. & Biol. (Cell Press). 21, 379-88.